Rare Variant Analysis Workflows: Analyzing NGS Data in Large Cohorts
- 1. Rare Variant Analysis Workflows: Analyzing NGS Data in Large Cohorts Nov 13, 2013 Bryce Christensen Statistical Geneticist / Director of Services
- 2. Rare Variant Analysis Workflows: Analyzing NGS Data in Large Cohorts Nov 13, 2013 Bryce Christensen Statistical Geneticist / Director of Services
- 3. Use the Questions pane in your GoToWebinar window Questions during the presentation
- 4. Golden Helix Leaders in Genetic Analytics Founded in 1998 Multi-disciplinary: computer science, bioinformatics, statistics, genetics Software and analytic services About Golden Helix
- 5. GenomeBrowse Free sequencing visualization tool Launched in 2011 Makes the process of exploring DNA- seq and RNA-seq pile-up and coverage data intuitive and powerful Stream public annotations via the cloud Use it to validate variant calls, trio exploration, de Novo discovery, and more
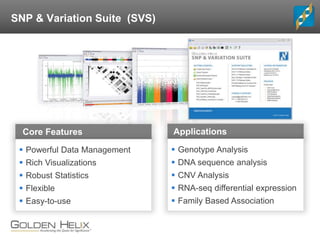
- 6. Core Features Packages Core Features Powerful Data Management Rich Visualizations Robust Statistics Flexible Easy-to-use Applications Genotype Analysis DNA sequence analysis CNV Analysis RNA-seq differential expression Family Based Association SNP & Variation Suite (SVS)
- 7. Merging of Two Great Products
- 8. Previous Webcast Recording Available
- 9. Agenda Brief review of upstream and QC considerations NGS workflow design in SVS 2 3 4 Overview of RV analysis approaches Define the problem: What is rare variant (RV) analysis?1 Interactive software demo5 What about exome chips?6 GenomeBrowse SVS 8: Exploratory tools, Analysis workflows
- 10. The Problem Array-based GWAS has been the primary technology for gene- finding research for most of the past decade - Common variant – common disease hypothesis NGS technology, particularly whole-exome sequencing, makes it possible to include rare variants (RVs) in the analysis Individual RVs lack statistical power for standard GWAS approaches - How do we utilize that information? Proposed solution: combine RVs into logical groups and analyze them as a single unit - AKA “Collapsing” or “Burden” tests.
- 11. From the Vault: January 2011 Slide on RV Analysis What have we learned since then?
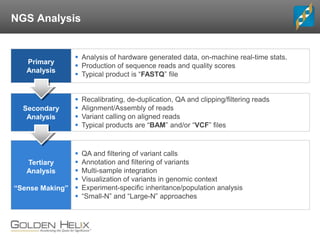
- 12. NGS Analysis Primary Analysis Secondary Analysis Tertiary Analysis “Sense Making” Analysis of hardware generated data, on-machine real-time stats. Production of sequence reads and quality scores Typical product is “FASTQ” file Recalibrating, de-duplication, QA and clipping/filtering reads Alignment/Assembly of reads Variant calling on aligned reads Typical products are “BAM” and/or “VCF” files QA and filtering of variant calls Annotation and filtering of variants Multi-sample integration Visualization of variants in genomic context Experiment-specific inheritance/population analysis “Small-N” and “Large-N” approaches
- 13. NGS Analysis Primary Analysis Secondary Analysis Tertiary Analysis “Sense Making” Analysis of hardware generated data, on-machine real-time stats. Production of sequence reads and quality scores Typical product is “FASTQ” file Recalibrating, de-duplication, QA and clipping/filtering reads Alignment/Assembly of reads Variant calling on aligned reads Typical products are “BAM” and/or “VCF” files QA and filtering of variant calls Annotation and filtering of variants Multi-sample integration Visualization of variants in genomic context Experiment-specific inheritance/population analysis “Small-N” and “Large-N” approaches
- 14. Most Importantly: Be Consistent! Gholson Lyon, 2012
- 15. Things That Can Confound Your Experiment Library preparation errors Sequencing errors Analysis errors PCR amplification point mutations (e.g. TruSeq protocol, amplicons) Emulsion PCR amplification point mutations (454, Ion Torrent and SOLiD) Bridge amplification errors (Illumina) Chimera generation (particularly during amplicon protocols) Sample contamination Amplification errors associated with high or low GC content PCR duplicates Base miscalls due to low signal InDel errors (particular PacBio) Short homopolymer associated InDels (Ion Torrent PGM) Post-homopolymeric tract SNPs (Illumina) and/or read-through problems Associated with inverted repeats (Illumina) Specific motifs particularly with older Illumina chemistry Calling variants without sufficient reads mapping Bad mapping (incorrectly placed read) Correctly placed read but InDels misaligned Multi-mapping to paralogous regions Sequence contamination e.g. adaptors Error in reference sequence Alignment to ends of contigs in draft assemblies Incorrect trimming of reads, aligning adaptors Inclusion of PCR duplicates Nick Loman: Sequencing data: I want the truth! (You can’t handle the truth!) Qual et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012 Jul
- 16. NGS Analysis Primary Analysis Secondary Analysis Tertiary Analysis “Sense Making” Analysis of hardware generated data, on-machine real-time stats. Production of sequence reads and quality scores Typical product is “FASTQ” file Recalibrating, de-duplication, QA and clipping/filtering reads Alignment/Assembly of reads Variant calling on aligned reads Typical products are “BAM” and/or “VCF” files QA and filtering of variant calls Annotation and filtering of variants Multi-sample integration Visualization of variants in genomic context Experiment-specific inheritance/population analysis “Small-N” and “Large-N” approaches
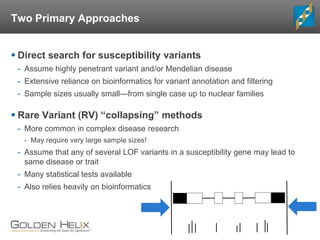
- 17. Two Primary Approaches Direct search for susceptibility variants - Assume highly penetrant variant and/or Mendelian disease - Extensive reliance on bioinformatics for variant annotation and filtering - Sample sizes usually small—from single case up to nuclear families Rare Variant (RV) “collapsing” methods - More common in complex disease research - May require very large sample sizes! - Assume that any of several LOF variants in a susceptibility gene may lead to same disease or trait - Many statistical tests available - Also relies heavily on bioinformatics
- 18. Families of Collapsing Tests Burden Tests - Combine minor alleles across multiple variant sites… - Without weighting (CMC, CAST, CMAT) - With fixed weights based on allele frequency (WSS, RWAS) - With data-adaptive weights (Lin/Tang, KBAC) - With data-adaptive thresholds (Step-Up, VT) - With extensions to allow for effects in either direction (Ionita-Laza/Lange, C-alpha) Kernel Tests - Allow for individual variant effects in either direction and permit covariate adjustment based on kernel regression - Kwee et al., AJHG, 2008 - SKAT - SKAT-O Credit: Schaid et al., Genet Epi, 2013
- 19. CMC: Combined Multivariate and Collapsing Multivariate test: simultaneous test for association of common and rare variants in gene Flexibility in variant frequency bin definition Testing methods include Hotelling T2 and Regression Regression method allows for covariate correction Li and Leal, AJHG, 2008
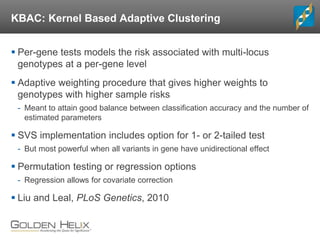
- 20. KBAC: Kernel Based Adaptive Clustering Per-gene tests models the risk associated with multi-locus genotypes at a per-gene level Adaptive weighting procedure that gives higher weights to genotypes with higher sample risks - Meant to attain good balance between classification accuracy and the number of estimated parameters SVS implementation includes option for 1- or 2-tailed test - But most powerful when all variants in gene have unidirectional effect Permutation testing or regression options - Regression allows for covariate correction Liu and Leal, PLoS Genetics, 2010
- 21. NGS Analysis Workflow Development in SVS SVS is very flexible in workflow design. SVS includes a broad range of tools for data manipulation and variant annotation and visualization that can be used together to guide us on an interactive exploration of the data. We begin by defining the final goal and the steps needed to help us reach that goal: - Are we looking for a very rare, non-synonymous variant that causes a dominant Mendelian trait? - Are we looking for a gene with excess rare variation in cases vs controls? Once we know what we are looking for, we can identify the available annotation sources that will help us answer the question.
- 22. Python Integration in SVS Allows rapid development and iteration of new functions API access to most SVS functions Access to extensive Python analytic libraries Fully documented in manual
- 23. SVS Online Scripts Repository Downloadable add-on functions for a variety of analysis and data management tasks “Plug-and-play” Some contributed by customers Popular scripts often get adopted into the “shipped” version of SVS. Scripts in repository are forward compatible to SVS 8.0
- 24. Today’s Featured Scripts Activate Variants by Genotype Count Threshold - Identify variants that occur with a specified frequency in one or several groups Filter by Marker Map Field - Variant-level “INFO” fields from VCF files are imported to the SVS marker map - This script allows you to filter markers based on those variables Many more useful scripts to take a look at: - Add Annotation Data to Marker Map from Spreadsheet - Nonparametric association tests - Import Unsorted VCF Files - Build Variant Spreadsheet - Many, many more
- 25. Interactive Demonstration GenomeBrowse - Exploring multi-sample VCF files in our free genome viewer software SVS 8.0 - Exploratory analysis workflow - Using downloaded scripts - Using basic analysis tools to create advanced workflows - Simulate the development of a burden test - RV association testing workflow - KBAC - CMC - Data visualization
- 26. SVS Demo
- 27. What about Exome Chips? Exome chips CAN be used with RV association tests Exome chips include both common and rare variants Remember: Exome chips don’t capture all rare variants. Exome chips are thus less powerful than WES for RV associations, but also significantly cheaper.
- 28. A Note about Exome Chips Exome chips are not GWAS chips - GWAS chips focus on common SNPs, have uniform spacing, minimal LD and are designed to capture population variability - Exome chips include rare variants and the content is anything but uniform Most GWAS functions can be used with exome chips, but require some workflow adjustments - Gender checking - IBD estimation - Principal components Not unlike other chips with custom/targeted content - Cardio-MetaboChip - ImmunoChip
- 29. Questions or more info: info@goldenhelix.com Request a copy of SVS at www.goldenhelix.com Download GenomeBrowse for free at www.GenomeBrowse.com
- 30. Use the Questions pane in your GoToWebinar window Any Questions?